R code
Example
library(magrittr)
library(data.table)
library(ggplot2)
library(ggpubr)
set.seed(123)
dt <-
data.table(`Body length` = seq(1, 5, 0.5)) %>%
.[, `Body weight` := 2 * `Body length` ^ (3 + rnorm(length(`Body length`), sd = 0.001))] %>%
rbind(., data.table(`Body length` = 6, `Body weight` = 200)) %>%
.[order(`Body length`), ]
dt.SMI <-
scaledMassIndex(dt$`Body length`, dt$`Body weight`, x.0 = 2)
summary(dt.SMI)
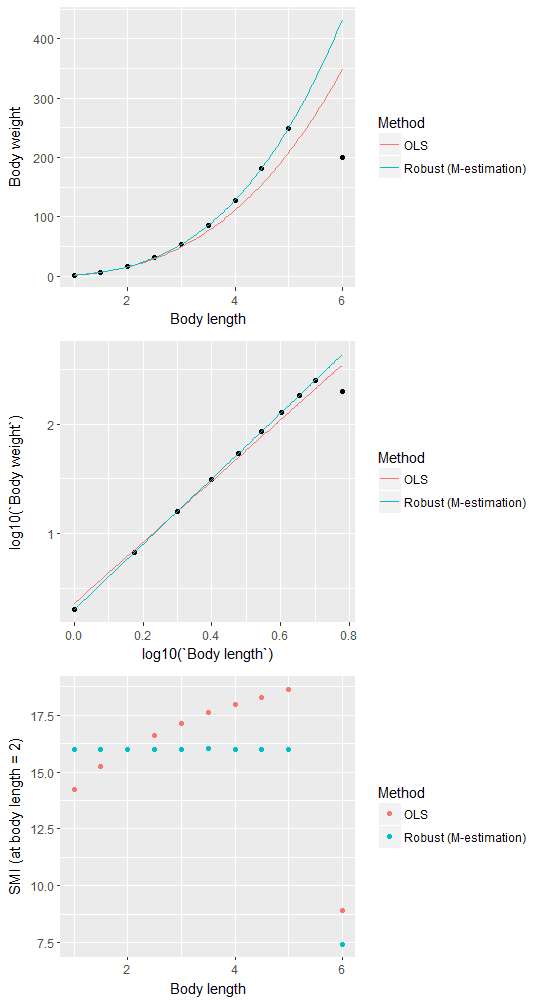
g1 <-
ggplot(dt, aes(`Body length`, `Body weight`)) +
geom_point() +
geom_line(aes(x, y, color = Method),
data = attr(dt.SMI, "pred") %>% melt(., "x", c("y.ols", "y.rob"), "Method", "y")) +
scale_colour_discrete(labels = c("OLS", "Robust (M-estimation)"))
g2 <-
ggplot(dt, aes(log10(`Body length`), log10(`Body weight`))) +
geom_point() +
geom_line(
aes(log10(x), log10(y), color = Method),
data = attr(dt.SMI, "pred") %>% melt(., "x", c("y.ols", "y.rob"), "Method", "y")
) +
scale_colour_discrete(labels = c("OLS", "Robust (M-estimation)"))
g3 <-
ggplot(melt(dt.SMI, "x", c("SMI.ols", "SMI.rob"), "Method"),
aes(x, value)) +
geom_point(aes(color = Method)) +
xlab("Body length") +
ylab("SMI (at body length = 2)") +
scale_colour_discrete(labels = c("OLS", "Robust (M-estimation)"))
ggarrange(g1,
g2,
g3,
ncol = 1,
nrow = 3,
align = "hv")

Thank you for sharing.
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteI could not found the function scaledMassIndex, used in the next code: scaledMassIndex(dt$`Body length`, dt$`Body weight`, x.0 = 2). Did i missunderstood the instructions or I have to install any package i do not have?
ReplyDeleteRun the whole function into R first, and then try the example.
DeleteI think there is some wrong specification in the plots. When running the example I cannot recreate the plots, as the data table has different names.
ReplyDeleteThis comment has been removed by the author.
DeleteI added attr(res, "pred") <- pred.DT to the function, and they should print
DeleteThanks and I have a neat give: How Many Houses Has Hometown Renovated renovate my house
ReplyDelete